Exposome Data Challenge Event
Powering the use of innovative methods for predicting health and disease in exposome studies
- Data
- 28/04/2021 - 30/04/2021
- Hora
- 2.30 PM CET (April 28) - 5.00 PM CET (April 30)
- Lloc
- Online
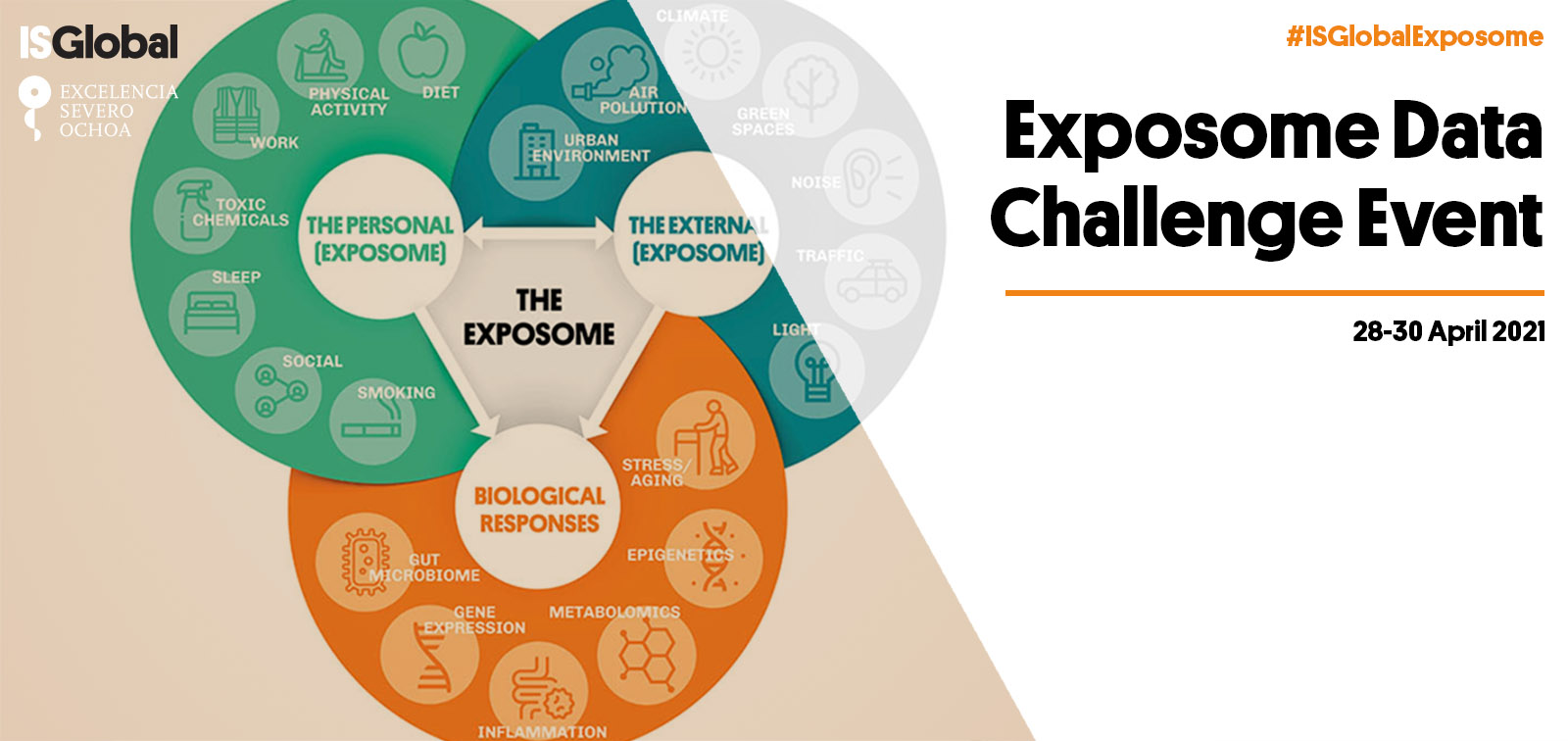
The exposome, described as "the totality of human environmental exposures from conception onwards", recognizes that individuals are exposed simultaneously to a multitude of different environmental factors and takes a holistic approach to the discovery of etiological factors for disease. The exposome’s main advantage over traditional ‘one-exposure-one-disease’ study approaches is that it provides an unprecedented conceptual framework for the study of multiple environmental hazards (urban, chemical, lifestyle, social) and their combined effects.
Click here to access the Agenda with videos and links to the presentation slides
Objective
The objective of this event was to promote innovative statistical, data science, or other quantitative approaches to studying the health effects of complex high-throughput measurement of exposure indicators (exposome). More detailed challenge examples were given on this document along with an overview of the dataset available. It brought together experts from the fields of environmental epidemiology and biostatistics from around the world including European partners from recently funded H2020 exposome projects (e.g. ATHLETE), fostering internal and external collaboration. This initiative was funded by the ISGlobal Severo Ochoa Programme, the ISGlobal Exposome Hub, and ATHLETE project.
In this working event, participants were offered an opportunity to test their statistical methods of choice on a real case scenario exposome dataset and later exhibit their findings at the workshop. The dataset included multiple health outcomes (continuous and categorical), multiple exposures, -omics and additional non-exposure variables (e.g., potential confounders).
The Exposome dataset represented a real case scenario of exposome dataset (based on the HELIX project database) with multiple correlated variables (N>100 exposure variables) arising from general and personal environments at different time points, biological molecular data (multi-omics: DNA methylation, gene expression, proteins, metabolomics) and multiple clinical phenotypes. The population was drawn from a multi-center study which represented the main confounding structure in the dataset.
A committee had assessed the abstracts. Based on these abstracts, subsets of individuals were invited to present their approach and statistical model(s) at the meeting. Young investigators were solicited as well as well established experts. Prizes and diplomas were awarded.
The workshop will result in a comprehensive document for publication that summarizes the findings from the workshop and outlines the most useful approaches and computational/conceptual/statistical models for determining or predicting health effects of high dimensional exposome datasets in collaboration with the event committee and the selected participants. We discussed advantages and disadvantages of different techniques.
Proposed Themes
- Exposome-health association studies
- Omics data integration in exposome-health studies (multi-omics, pathway analysis, mediation…)
- Causal inference techniques (inferring/validating causal structure from high-dimensional heterogeneous data, techniques to obtain causal estimates with high-dimensional data…)
- High-dimensional data mediation analysis
- Mixture, combined or “cocktail” effects
- Hierarchical modelling
- Non-linear effects and high-order interactions
- Machine learning techniques
- Data visualization/summary measures (new plots, new metrics…)
The approaches may or may not try to accommodate some of the particularities of the data (multi-cohort, missing data, count responses, categorical and continuous variables…). Add More detailed challenge examples are given on this document along with an overview of the dataset available.
Event Committee
Léa Maitre, ISGlobal, Spain (Exposome, omics)
Xavier Basagaña, ISGlobal, Spain (Biostatistics)
Juan Ramón González, ISGlobal, Spain (Bioinformatics)
Charline Warembourg, ISGlobal/INSERM, Spain/France (Biostatistics)
Paula Petrone, ISGlobal, Spain (Biomedical Data Science)
Marc Chadeau, Imperial College London (Statistics, omics, exposome)
More Information
E-mail: rodney.ortiz@isglobal.org
Paper following the the Exposome Data Challenge
Léa Maitre, Jean-Baptiste Guimbaud, Charline Warembourg, Nuria Güil-Oumrait, Paula Marcela Petrone, Marc Chadeau-Hyam, Martine Vrijheid, Xavier Basagaña, Juan R. Gonzalez, State-of-the-art methods for exposure-health studies: Results from the exposome data challenge event, Environment International, Volume 168, 2022, 107422, ISSN 0160-4120, https://doi.org/10.1016/j.envint.2022.107422.
(https://www.sciencedirect.com/science/article/pii/S016041202200349X)
We acknowledge support from the Spanish Ministry of Science and Innovation through the “Centro de Excelencia Severo Ochoa 2019-2023” Program (CEX2018-000806-S), and support from the Generalitat de Catalunya through the CERCA Program. ATHLETE project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 874583.



